Pessimism of the Intellect, Optimism of the Will Favorite posts | Manifold podcast | Twitter: @hsu_steve
Friday, July 27, 2018
Insight Podcast: James Lee interview on SSGAC EA3
Spencer Wells and Razib Khan interview James Lee (Professor of Psychology, University of Minnesota, BA Berkeley, PhD Harvard) about the recent SSGAC EA3 GWAS.
Comment: James mentions that EA3 may be approaching the GCTA h2 limit (~0.15? so limiting r ~ 0.4) already. But the limit for actual cognitive ability is much higher; with enough data I think we could get to r ~ 0.6 or even r ~ 0.7 eventually for common SNPs -- similar to height.
United Club, HK International Airport
James, me, Chris Chang. (About $1M worth of Illumina HiSeqs in crates behind us?)
Wednesday, July 25, 2018
Genomic Prediction: A Hypothetical (Embryo Selection)
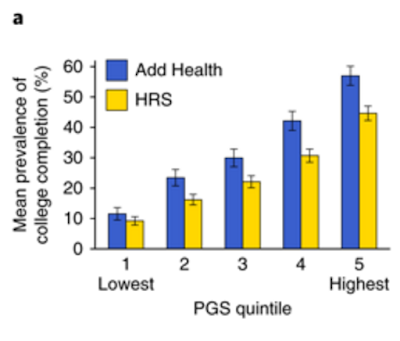
The new SSGAC EA3 paper in Nature Genetics contains the following figure.
Add Health (National Longitudinal Study of Adolescent to Adult Health) and HRS (Health in Retirement Study) are two longitudinal cohorts under study by social scientists. Horizontal axis is polygenic score (computed from DNA alone). It appears that individuals with top quintile polygenic scores are about 5 times more likely to complete college than bottom quintile individuals. (IIUC, HRS cohort grew up in an earlier era when college attendance rates were lower; Add Health participants are younger.)
Consider the following hypothetical:
See Nativity 2050 and The Future is Here: Genomic Prediction in MIT Technology Review.
Add Health (National Longitudinal Study of Adolescent to Adult Health) and HRS (Health in Retirement Study) are two longitudinal cohorts under study by social scientists. Horizontal axis is polygenic score (computed from DNA alone). It appears that individuals with top quintile polygenic scores are about 5 times more likely to complete college than bottom quintile individuals. (IIUC, HRS cohort grew up in an earlier era when college attendance rates were lower; Add Health participants are younger.)
Consider the following hypothetical:
You are an IVF physician advising parents who have exactly 2 viable embryos, ready for implantation. The parents want to implant only one embryo.
All genetic and morphological information about the embryos suggest that they are both viable, healthy, and free of elevated disease risk.Note, in the very near future this question will no longer be hypothetical...
However, embryo A has polygenic score (as in figure above) in the lowest quintile (elevated risk of struggling in school) while embryo B has polygenic score in the highest quintile (less than average risk of struggling in school). We could sharpen the question by assuming, e.g., that embryo A has score in the bottom 1% while embryo B is in the top 1%.
You have no other statistical or medical information to differentiate between the two embryos.
What do you tell the parents? Do you inform them about the polygenic score difference between the embryos?
See Nativity 2050 and The Future is Here: Genomic Prediction in MIT Technology Review.
Monday, July 23, 2018
SSGAC EA3: genomic prediction of educational attainment and related cognitive phenotypes
Years ago I predicted that:
1. Cognitive ability would turn out to be influenced by many thousands of genetic variants, each of small effect.
2. With large enough sample size we would detect these variants and eventually construct genomic predictors.
The Nature Genetics paper below from the SSGAC collaboration takes a significant step in that direction.
Although the study used over a million genotypes, the data had to be aggregated across many sub-cohorts using summary statistics only. This does not permit the L1-penalized optimization we used to build our height predictor.
For out of sample validation of the results below, see this PNAS paper, which (unusually) appeared before the paper on which it is based.
The lead author James Lee is on the left below. Chris Chang, author of Plink 2.0, is on the right. The photo was taken in 2010 at BGI -- they are standing in front of crates of Illumina sequencers.

Here's a comment on the paper I provided to a journalist:
1. Cognitive ability would turn out to be influenced by many thousands of genetic variants, each of small effect.
2. With large enough sample size we would detect these variants and eventually construct genomic predictors.
The Nature Genetics paper below from the SSGAC collaboration takes a significant step in that direction.
Although the study used over a million genotypes, the data had to be aggregated across many sub-cohorts using summary statistics only. This does not permit the L1-penalized optimization we used to build our height predictor.
For out of sample validation of the results below, see this PNAS paper, which (unusually) appeared before the paper on which it is based.
The lead author James Lee is on the left below. Chris Chang, author of Plink 2.0, is on the right. The photo was taken in 2010 at BGI -- they are standing in front of crates of Illumina sequencers.

Article | Published: 23 July 2018A nice figure from the paper: Add Health (National Longitudinal Study of Adolescent to Adult Health) and HRS (Health in Retirement Study) are two longitudinal cohorts that have been genotyped; horizontal axis is polygenic score. It appears that individuals with top quintile polygenic scores are about 5 times more likely to complete college than bottom quintile individuals.
Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals
James J. Lee, Robbee Wedow, […]David Cesarini
Nature Genetics (2018)
Abstract
Here we conducted a large-scale genetic association analysis of educational attainment in a sample of approximately 1.1 million individuals and identify 1,271 independent genome-wide-significant SNPs. For the SNPs taken together, we found evidence of heterogeneous effects across environments. The SNPs implicate genes involved in brain-development processes and neuron-to-neuron communication. In a separate analysis of the X chromosome, we identify 10 independent genome-wide-significant SNPs and estimate a SNP heritability of around 0.3% in both men and women, consistent with partial dosage compensation. A joint (multi-phenotype) analysis of educational attainment and three related cognitive phenotypes generates polygenic scores that explain 11–13% of the variance in educational attainment and 7–10% of the variance in cognitive performance. This prediction accuracy substantially increases the utility of polygenic scores as tools in research.
Here's a comment on the paper I provided to a journalist:
The EA3 predictor correlates about 0.35 with educational attainment, and slightly less well with measured cognitive ability. While this is far from perfect prediction, it does allow identification of individuals, using DNA alone, who are at unusual risk of being well below average in cognitive ability or struggling in school. Standardized tests, such as SAT, ACT, GRE, LSAT, etc., typically also correlate roughly 0.35 with educational outcomes like grade point average, degree completion, etc. In this sense, the genomic predictor is comparable to widely used tests and it will certainly improve as more data are analyzed. See figure.
Sunday, July 22, 2018
LATTICE 2018 at MSU
The 36th Annual International Symposium on Lattice Field Theory begins tomorrow, hosted by MSU. My opening remarks are below. No peeking if you are an attendee!
LATTICE 2018 Opening Remarks 7/23/2018
Good morning. I’d like to extend my warmest welcome to all of you on behalf of Michigan State University. We are very pleased and honored to be the hosts for The 36th Annual International Symposium on Lattice Field Theory.
It is my opinion that even within Physics, and even within Theoretical Physics, Lattice Field Theory is underappreciated. The idea that we can constructively realize quantum field theories in silico, that we can perform precision calculations in the deepest models of fundamental physics, is really incredible. It has taken many decades to get to this point: to master strongly coupled quantum fluctuations, spacetime trajectories of quantum fields like quarks and gluons, advanced algorithms and hardware designs, matching to effective field theories, and many other conceptually beautiful but ultimately concrete things.
Along with some recent AI advances like AlphaGo, the precise ab initio calculation of physical quantities in lattice QCD must be considered among the most impressive computations performed by the human species. If some Alien visitors were evaluating the accomplishments of our civilization, I would want them to take into account the work of people here today.
I first became aware of lattice gauge theory from John Preskill’s lecture notes for Physics 234, a year-long Caltech course on advanced topics in QCD. I never imagined, back in the 1980s, the successes that all of you have achieved today. The important message to young people is that one should not be dissuaded from attempting difficult projects.
At MSU we made the decision a few years ago to invest in lattice physics. We went from no lattice researchers, to one of the larger groups in the US. One of the drivers for this decision was the hope that lattice simulations would one day connect QCD to the experimental results coming from FRIB -- the MSU / DOE Facility for Rare Isotope Beams. Today we can compute, from first principles, the properties of light hadrons. In the coming decades, I believe we will compute real time scattering amplitudes and nuclear forces from QCD itself.
DOE and MSU are investing, all told, roughly a billion dollars in FRIB. While it is the Experimentalists who build and run the machine, and deserve the main credit, we as Theorists have the responsibility to ensure that the results of the experiment inform our deeper understanding of nuclear physics and QCD. Physicists are not stamp collectors -- we do not measure things just to measure them. We measure things which are important and have deep implications.
To reach the long awaited goal of connecting nuclear physics directly to QCD, we depend on the lattice community, on all of you. May the next 30 years see as much progress as the last.
Thank you very much.
Action photos!
London Calling
On my way home from Stockholm / ICML I stopped in London to see my friend Dominic Cummings, give a talk at ASI Data Science, and have some Oligarch meetings. Sorry I can't share more details.
Here are some photos from the British Museum.
Bodhisattva: a person who is able to reach nirvana but delays doing so out of compassion in order to save suffering beings.
Here are some photos from the British Museum.
Bodhisattva: a person who is able to reach nirvana but delays doing so out of compassion in order to save suffering beings.
“Tenfold be your damnation," he said.. "There shall be no rebirth."
His hands came open then. A tall, nobly proportioned man lay upon the floor at his feet, his head resting upon his right shoulder.
His eye had finally closed.
Yama turned the corpse with the toe of his boot. "Build a pyre and burn this body," he said to the monks, not turning toward them. "Spare none of the rites. One of the highest has died this day.”
Lord of Light, Roger Zelazny.
Tuesday, July 17, 2018
ICML notes
It's never been a better time to work on AI/ML. Vast resources are being deployed in this direction, by corporations and governments alike. In addition to the marvelous practical applications in development, a theoretical understanding of Deep Learning may emerge in the next few years.
The notes below are to keep track of some interesting things I encountered at the meeting.
Some ML learning resources:
Metacademy
Depth First study of AlphaGo
I heard a more polished version of this talk by Elad at the Theory of Deep Learning workshop. He is trying to connect results in sparse learning (e.g., performance guarantees for L1 or threshold algos) to Deep Learning. (Video is from UCLA IPAM.)
It may turn out that the problems on which DL works well are precisely those in which the training data (and underlying generative processes) have a hierarchical structure which is sparse, level by level. Layered networks perform a kind of coarse graining (renormalization group flow): first layers filter by feature, subsequent layers by combinations of features, etc. But the whole thing can be understood as products of sparse filters, and the performance under training is described by sparse performance guarantees (ReLU = thresholded penalization?). Given the inherent locality of physics (atoms, molecules, cells, tissue; atoms, words, sentences, ...) it is not surprising that natural phenomena generate data with this kind of hierarchical structure.
Off-topic: At dinner with one of my former students and his colleague (both researchers at an AI lab in Germany), the subject of Finitism came up due to a throwaway remark about the Continuum Hypothesis.
Wikipedia
Horizons of Truth
Chaitin on Physics and Mathematics
David Deutsch:
Paul Cohen:
The notes below are to keep track of some interesting things I encountered at the meeting.
Some ML learning resources:
Metacademy
Depth First study of AlphaGo
I heard a more polished version of this talk by Elad at the Theory of Deep Learning workshop. He is trying to connect results in sparse learning (e.g., performance guarantees for L1 or threshold algos) to Deep Learning. (Video is from UCLA IPAM.)
It may turn out that the problems on which DL works well are precisely those in which the training data (and underlying generative processes) have a hierarchical structure which is sparse, level by level. Layered networks perform a kind of coarse graining (renormalization group flow): first layers filter by feature, subsequent layers by combinations of features, etc. But the whole thing can be understood as products of sparse filters, and the performance under training is described by sparse performance guarantees (ReLU = thresholded penalization?). Given the inherent locality of physics (atoms, molecules, cells, tissue; atoms, words, sentences, ...) it is not surprising that natural phenomena generate data with this kind of hierarchical structure.
Off-topic: At dinner with one of my former students and his colleague (both researchers at an AI lab in Germany), the subject of Finitism came up due to a throwaway remark about the Continuum Hypothesis.
Wikipedia
Horizons of Truth
Chaitin on Physics and Mathematics
David Deutsch:
The reason why we find it possible to construct, say, electronic calculators, and indeed why we can perform mental arithmetic, cannot be found in mathematics or logic. The reason is that the laws of physics "happen" to permit the existence of physical models for the operations of arithmetic such as addition, subtraction and multiplication.My perspective: We experience the physical world directly, so the highest confidence belief we have is in its reality. Mathematics is an invention of our brains, and cannot help but be inspired by the objects we find in the physical world. Our idealizations (such as "infinity") may or may not be well-founded. In fact, mathematics with infinity included may be very sick, as evidenced by Godel's results, or paradoxes in set theory. There is no reason that infinity is needed (as far as we know) to do physics. It is entirely possible that there are only a (large but) finite number of degrees of freedom in the physical universe.
Paul Cohen:
I will ascribe to Skolem a view, not explicitly stated by him, that there is a reality to mathematics, but axioms cannot describe it. Indeed one goes further and says that there is no reason to think that any axiom system can adequately describe it.This "it" (mathematics) that Cohen describes may be the set of idealizations constructed by our brains extrapolating from physical reality. But there is no guarantee that these idealizations have a strong kind of internal consistency and indeed they cannot be adequately described by any axiom system.
Sunday, July 15, 2018
Monday, July 09, 2018
Game Over: Genomic Prediction of Social Mobility
[ NOTE: The PNAS paper discussed below uses the SSGAC EA3 genomic predictor, trained on over a million genomes. The EA3 paper has now appeared in Nature Genetics. ]
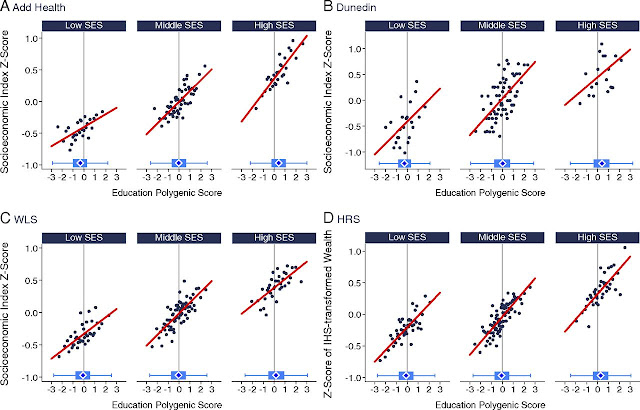
The figure below shows SNP-based polygenic score and life outcome (socioeconomic index, on vertical axis) in four longitudinal cohorts, one from New Zealand (Dunedin) and three from the US. Each cohort (varying somewhat in size) has thousands of individuals, ~20k in total (all of European ancestry). The points displayed are averages over bins containing 10-50 individuals. For each cohort, the individuals have been grouped by childhood (family) social economic status. Social mobility can be predicted from polygenic score. Note that higher SES families tend to have higher polygenic scores on average -- which is what one might expect from a society that is at least somewhat meritocratic. The cohorts have not been used in training -- this is true out-of-sample validation. Furthermore, the four cohorts represent different geographic regions (even, different continents) and individuals born in different decades.
Everyone should stop for a moment and think carefully about the implications of the paragraph above and the figure below.
Caption from the PNAS paper.
The paper:
Note Added from comments: Plots would look much noisier if not for averaging many individuals into single point. Keep in mind that socioeconomic success depends on a lot more than just cognitive ability, or even cognitive ability + conscientiousness.
But, underlying predictor correlates ~0.35 with actual educational attainment, IIRC. That is, the polygenic score predicts EA about as well as standardized tests predict success in schooling.
This means you can at least use it to identify outliers: just as a very high/low test score (SAT, ACT, GRE) does not *guarantee* success/failure in school, nevertheless the signal is useful for selection = admissions.
The figure below shows SNP-based polygenic score and life outcome (socioeconomic index, on vertical axis) in four longitudinal cohorts, one from New Zealand (Dunedin) and three from the US. Each cohort (varying somewhat in size) has thousands of individuals, ~20k in total (all of European ancestry). The points displayed are averages over bins containing 10-50 individuals. For each cohort, the individuals have been grouped by childhood (family) social economic status. Social mobility can be predicted from polygenic score. Note that higher SES families tend to have higher polygenic scores on average -- which is what one might expect from a society that is at least somewhat meritocratic. The cohorts have not been used in training -- this is true out-of-sample validation. Furthermore, the four cohorts represent different geographic regions (even, different continents) and individuals born in different decades.
Everyone should stop for a moment and think carefully about the implications of the paragraph above and the figure below.
Caption from the PNAS paper.
Fig. 4. Education polygenic score associations with social attainment for Add Health Study, WLS, Dunedin Study, and HRS participants with low-, middle-, and high-socioeconomic status (SES) social origins. The figure plots polygenic score associations with socioeconomic attainment for Add Health Study (A), Dunedin Study (B), WLS (C), and HRS (D) participants who grew up in low-, middle-, and high-SES households. For the figure, low- middle-, and high-SES households were defined as the bottom quartile, middle 50%, and top quartile of the social origins score distributions for the Add Health Study, WLS, and HRS. For the Dunedin Study, low SES was defined as a childhood NZSEI of two or lower (20% of the sample), middle SES was defined as childhood NZSEI of three to four (63% of the sample), and high SES was defined as childhood NZSEI of five or six (17% of the sample). Attainment is graphed in terms of socioeconomic index scores for the Add Health Study, Dunedin Study, and WLS and in terms of household wealth in the HRS. Add Health Study and WLS socioeconomic index scores were calculated from Hauser and Warren (34) occupational income and occupational education scores. Dunedin Study socioeconomic index scores were calculated similarly, according to the Statistics New Zealand NZSEI (38). HRS household wealth was measured from structured interviews about assets. All measures were z-transformed to have mean = 0, SD = 1 for analysis. The individual graphs show binned scatterplots in which each plotted point reflects average x and y coordinates for a bin of 50 participants for the Add Health Study, WLS, and HRS and for a bin of 10 participants for the Dunedin Study. The red regression lines are plotted from the raw data. The box-and-whisker plots at the bottom of the graphs show the distribution of the education polygenic score for each childhood SES category. The blue diamond in the middle of the box shows the median; the box shows the interquartile range; and the whiskers show upper and lower bounds defined by the 25th percentile minus 1.5× the interquartile range and the 75th percentile plus 1.5× the interquartile range, respectively. The vertical line intersecting the x axis shows the cohort average polygenic score. The figure illustrates three findings observed consistently across cohorts: (i) participants who grew up in higher-SES households tended to have higher socioeconomic attainment independent of their genetics compared with peers who grew up in lower-SES households; (ii) participants’ polygenic scores were correlated with their social origins such that those who grew up in higher-SES households tended to have higher polygenic scores compared with peers who grew up in lower-SES households; (iii) participants with higher polygenic scores tended to achieve higher levels of attainment across strata of social origins, including those born into low-SES families.
The paper:
Genetic analysis of social-class mobility in five longitudinal studies, Belsky et al.
PNAS July 9, 2018. 201801238; published ahead of print July 9, 2018. https://doi.org/10.1073/pnas.1801238115
A summary genetic measure, called a “polygenic score,” derived from a genome-wide association study (GWAS) of education can modestly predict a person’s educational and economic success. This prediction could signal a biological mechanism: Education-linked genetics could encode characteristics that help people get ahead in life. Alternatively, prediction could reflect social history: People from well-off families might stay well-off for social reasons, and these families might also look alike genetically. A key test to distinguish biological mechanism from social history is if people with higher education polygenic scores tend to climb the social ladder beyond their parents’ position. Upward mobility would indicate education-linked genetics encodes characteristics that foster success. We tested if education-linked polygenic scores predicted social mobility in >20,000 individuals in five longitudinal studies in the United States, Britain, and New Zealand. Participants with higher polygenic scores achieved more education and career success and accumulated more wealth. However, they also tended to come from better-off families. In the key test, participants with higher polygenic scores tended to be upwardly mobile compared with their parents. Moreover, in sibling-difference analysis, the sibling with the higher polygenic score was more upwardly mobile. Thus, education GWAS discoveries are not mere correlates of privilege; they influence social mobility within a life. Additional analyses revealed that a mother’s polygenic score predicted her child’s attainment over and above the child’s own polygenic score, suggesting parents’ genetics can also affect their children’s attainment through environmental pathways. Education GWAS discoveries affect socioeconomic attainment through influence on individuals’ family-of-origin environments and their social mobility.
Note Added from comments: Plots would look much noisier if not for averaging many individuals into single point. Keep in mind that socioeconomic success depends on a lot more than just cognitive ability, or even cognitive ability + conscientiousness.
But, underlying predictor correlates ~0.35 with actual educational attainment, IIRC. That is, the polygenic score predicts EA about as well as standardized tests predict success in schooling.
This means you can at least use it to identify outliers: just as a very high/low test score (SAT, ACT, GRE) does not *guarantee* success/failure in school, nevertheless the signal is useful for selection = admissions.
Friday, July 06, 2018
Seven Years, Two Tweets
Is anyone keeping score?
See On the Genetic Architecture of Cognitive Ability (2014) and Nautilus Magazine: Super Intelligent Humans.
See On the Genetic Architecture of Cognitive Ability (2014) and Nautilus Magazine: Super Intelligent Humans.
This 2011 Google Talk with @hsu_steve changed my life more than any book, political movement, or event https://t.co/SXCmoGMqNV— Nishiki (@NishikiPrestige) July 5, 2018
2011: we sought *first* genetic variant correlated to g at genome wide statistical significance. 2018: found thousands of such SNPs and can crudely predict g from genotype alone. 2011 predictions: additive effects, highly polygenic architecture, large samples, etc. all came true!— steve hsu (@hsu_steve) July 6, 2018
Thursday, July 05, 2018
Cognitive ability predicted from fMRI (Caltech Neuroscience)
Caltech researchers used elastic net (L1 and L2 penalization) to train a predictor using cognitive scores and fMRI data from ~900 individuals. The predictor captures about 20% of variance in intelligence; the score correlates a bit more than 0.45 with actual intelligence. This may validate earlier work by Korean researchers in 2015, although the Korean group claimed much higher predictive correlations.
Press release:
Press release:
In a new study, researchers from Caltech, Cedars-Sinai Medical Center, and the University of Salerno show that their new computing tool can predict a person's intelligence from functional magnetic resonance imaging (fMRI) scans of their resting state brain activity. Functional MRI develops a map of brain activity by detecting changes in blood flow to specific brain regions. In other words, an individual's intelligence can be gleaned from patterns of activity in their brain when they're not doing or thinking anything in particular—no math problems, no vocabulary quizzes, no puzzles.Paper:
"We found if we just have people lie in the scanner and do nothing while we measure the pattern of activity in their brain, we can use the data to predict their intelligence," says Ralph Adolphs (PhD '92), Bren Professor of Psychology, Neuroscience, and Biology, and director and Allen V. C. Davis and Lenabelle Davis Leadership Chair of the Caltech Brain Imaging Center.
To train their algorithm on the complex patterns of activity in the human brain, Adolphs and his team used data collected by the Human Connectome Project (HCP), a scientific endeavor funded by the National Institutes of Health (NIH) that seeks to improve understanding of the many connections in the human brain. Adolphs and his colleagues downloaded the brain scans and intelligence scores from almost 900 individuals who had participated in the HCP, fed these into their algorithm, and set it to work.
After processing the data, the team's algorithm was able to predict intelligence at statistically significant levels across these 900 subjects, says Julien Dubois (PhD '13), a postdoctoral fellow at Cedars-Sinai Medical Center. But there is a lot of room for improvement, he adds. The scans are coarse and noisy measures of what is actually happening in the brain, and a lot of potentially useful information is still being discarded.
"The information that we derive from the brain measurements can be used to account for about 20 percent of the variance in intelligence we observed in our subjects," Dubois says. "We are doing very well, but we are still quite far from being able to match the results of hour-long intelligence tests, like the Wechsler Adult Intelligence Scale,"
Dubois also points out a sort of philosophical conundrum inherent in the work. "Since the algorithm is trained on intelligence scores to begin with, how do we know that the intelligence scores are correct?" The researchers addressed this issue by extracting a more precise estimate of intelligence across 10 different cognitive tasks that the subjects had taken, not only from an IQ test. ...
A distributed brain network predicts general intelligence from resting-state human neuroimaging data
Individual people differ in their ability to reason, solve problems, think abstractly, plan and learn. A reliable measure of this general ability, also known as intelligence, can be derived from scores across a diverse set of cognitive tasks. There is great interest in understanding the neural underpinnings of individual differences in intelligence, since it is the single best predictor of long-term life success, and since individual differences in a similar broad ability are found across animal species. The most replicated neural correlate of human intelligence to date is total brain volume. However, this coarse morphometric correlate gives no insights into mechanisms; it says little about function. Here we ask whether measurements of the activity of the resting brain (resting-state fMRI) might also carry information about intelligence. We used the final release of the Young Adult Human Connectome Project dataset (N=884 subjects after exclusions), providing a full hour of resting-state fMRI per subject; controlled for gender, age, and brain volume; and derived a reliable estimate of general intelligence from scores on multiple cognitive tasks. Using a cross-validated predictive framework, we predicted 20% of the variance in general intelligence in the sampled population from their resting-state fMRI data. Interestingly, no single anatomical structure or network was responsible or necessary for this prediction, which instead relied on redundant information distributed across the brain.
Tuesday, July 03, 2018
In the land of the Gene Titans
Apologies for the lack of posts recently. I've been traveling and busy with meetings. For my own recollection, here is a partial list of places I've been in the past weeks.
Illumina (San Diego)
Ancestry (~10M genomes! San Francisco)
23andMe (~5M genomes! Mountain View)
OpenAI (machines beat pro human teams in complex Dota 2 game! San Francisco)
Affymetrix (Santa Clara)
Healdsburg, Sonoma (Talk at meeting of Oligarchs :-)
Soros Fund Management (Talk at leadership retreat, Museum of Arts and Design, NYC)
These GeneTitans are part of the Affy lab that did all of the genotyping for the UK Biobank project. The footprint for this kind of lab is shockingly small: ~6k samples per week per machine and ~10 machines means millions of individual genotypes per year. Illumina produces similar arrays/readers and a hundred square meters of lab space is enough to process millions of samples per year for DTC genomics companies like 23andMe and Ancestry.
We may have a lab like this soon at MSU ;-)
Illumina (San Diego)
Ancestry (~10M genomes! San Francisco)
23andMe (~5M genomes! Mountain View)
OpenAI (machines beat pro human teams in complex Dota 2 game! San Francisco)
Affymetrix (Santa Clara)
Healdsburg, Sonoma (Talk at meeting of Oligarchs :-)
Soros Fund Management (Talk at leadership retreat, Museum of Arts and Design, NYC)
These GeneTitans are part of the Affy lab that did all of the genotyping for the UK Biobank project. The footprint for this kind of lab is shockingly small: ~6k samples per week per machine and ~10 machines means millions of individual genotypes per year. Illumina produces similar arrays/readers and a hundred square meters of lab space is enough to process millions of samples per year for DTC genomics companies like 23andMe and Ancestry.
We may have a lab like this soon at MSU ;-)
Subscribe to:
Posts (Atom)
Blog Archive
-
▼
2018
(128)
-
▼
07
(11)
- Insight Podcast: James Lee interview on SSGAC EA3
- Genomic Prediction: A Hypothetical (Embryo Selection)
- SSGAC EA3: genomic prediction of educational attai...
- LATTICE 2018 at MSU
- London Calling
- ICML notes
- AI Lollapalooza in Stockholm
- Game Over: Genomic Prediction of Social Mobility
- Seven Years, Two Tweets
- Cognitive ability predicted from fMRI (Caltech Neu...
- In the land of the Gene Titans
-
▼
07
(11)
Labels
- physics (420)
- genetics (325)
- globalization (301)
- genomics (295)
- technology (282)
- brainpower (280)
- finance (275)
- american society (261)
- China (249)
- innovation (231)
- ai (206)
- economics (202)
- psychometrics (190)
- science (172)
- psychology (169)
- machine learning (166)
- biology (163)
- photos (162)
- genetic engineering (150)
- universities (150)
- travel (144)
- podcasts (143)
- higher education (141)
- startups (139)
- human capital (127)
- geopolitics (124)
- credit crisis (115)
- political correctness (108)
- iq (107)
- quantum mechanics (107)
- cognitive science (103)
- autobiographical (97)
- politics (93)
- careers (90)
- bounded rationality (88)
- social science (86)
- history of science (85)
- realpolitik (85)
- statistics (83)
- elitism (81)
- talks (80)
- evolution (79)
- credit crunch (78)
- biotech (76)
- genius (76)
- gilded age (73)
- income inequality (73)
- caltech (68)
- books (64)
- academia (62)
- history (61)
- intellectual history (61)
- MSU (60)
- sci fi (60)
- harvard (58)
- silicon valley (58)
- mma (57)
- mathematics (55)
- education (53)
- video (52)
- kids (51)
- bgi (48)
- black holes (48)
- cdo (45)
- derivatives (43)
- neuroscience (43)
- affirmative action (42)
- behavioral economics (42)
- economic history (42)
- literature (42)
- nuclear weapons (42)
- computing (41)
- jiujitsu (41)
- physical training (40)
- film (39)
- many worlds (39)
- quantum field theory (39)
- expert prediction (37)
- ufc (37)
- bjj (36)
- bubbles (36)
- mortgages (36)
- google (35)
- race relations (35)
- hedge funds (34)
- security (34)
- von Neumann (34)
- meritocracy (31)
- feynman (30)
- quants (30)
- taiwan (30)
- efficient markets (29)
- foo camp (29)
- movies (29)
- sports (29)
- music (28)
- singularity (27)
- entrepreneurs (26)
- conferences (25)
- housing (25)
- obama (25)
- subprime (25)
- venture capital (25)
- berkeley (24)
- epidemics (24)
- war (24)
- wall street (23)
- athletics (22)
- russia (22)
- ultimate fighting (22)
- cds (20)
- internet (20)
- new yorker (20)
- blogging (19)
- japan (19)
- scifoo (19)
- christmas (18)
- dna (18)
- gender (18)
- goldman sachs (18)
- university of oregon (18)
- cold war (17)
- cryptography (17)
- freeman dyson (17)
- smpy (17)
- treasury bailout (17)
- algorithms (16)
- autism (16)
- personality (16)
- privacy (16)
- Fermi problems (15)
- cosmology (15)
- happiness (15)
- height (15)
- india (15)
- oppenheimer (15)
- probability (15)
- social networks (15)
- wwii (15)
- fitness (14)
- government (14)
- les grandes ecoles (14)
- neanderthals (14)
- quantum computers (14)
- blade runner (13)
- chess (13)
- hedonic treadmill (13)
- nsa (13)
- philosophy of mind (13)
- research (13)
- aspergers (12)
- climate change (12)
- harvard society of fellows (12)
- malcolm gladwell (12)
- net worth (12)
- nobel prize (12)
- pseudoscience (12)
- Einstein (11)
- art (11)
- democracy (11)
- entropy (11)
- geeks (11)
- string theory (11)
- television (11)
- Go (10)
- ability (10)
- complexity (10)
- dating (10)
- energy (10)
- football (10)
- france (10)
- italy (10)
- mutants (10)
- nerds (10)
- olympics (10)
- pop culture (10)
- crossfit (9)
- encryption (9)
- eugene (9)
- flynn effect (9)
- james salter (9)
- simulation (9)
- tail risk (9)
- turing test (9)
- alan turing (8)
- alpha (8)
- ashkenazim (8)
- data mining (8)
- determinism (8)
- environmentalism (8)
- games (8)
- keynes (8)
- manhattan (8)
- new york times (8)
- pca (8)
- philip k. dick (8)
- qcd (8)
- real estate (8)
- robot genius (8)
- success (8)
- usain bolt (8)
- Iran (7)
- aig (7)
- basketball (7)
- free will (7)
- fx (7)
- game theory (7)
- hugh everett (7)
- inequality (7)
- information theory (7)
- iraq war (7)
- markets (7)
- paris (7)
- patents (7)
- poker (7)
- teaching (7)
- vietnam war (7)
- volatility (7)
- anthropic principle (6)
- bayes (6)
- class (6)
- drones (6)
- econtalk (6)
- empire (6)
- global warming (6)
- godel (6)
- intellectual property (6)
- nassim taleb (6)
- noam chomsky (6)
- prostitution (6)
- rationality (6)
- academia sinica (5)
- bobby fischer (5)
- demographics (5)
- fake alpha (5)
- kasparov (5)
- luck (5)
- nonlinearity (5)
- perimeter institute (5)
- renaissance technologies (5)
- sad but true (5)
- software development (5)
- solar energy (5)
- warren buffet (5)
- 100m (4)
- Poincare (4)
- assortative mating (4)
- bill gates (4)
- borges (4)
- cambridge uk (4)
- censorship (4)
- charles darwin (4)
- computers (4)
- creativity (4)
- hormones (4)
- humor (4)
- judo (4)
- kerviel (4)
- microsoft (4)
- mixed martial arts (4)
- monsters (4)
- moore's law (4)
- soros (4)
- supercomputers (4)
- trento (4)
- 200m (3)
- babies (3)
- brain drain (3)
- charlie munger (3)
- cheng ting hsu (3)
- chet baker (3)
- correlation (3)
- ecosystems (3)
- equity risk premium (3)
- facebook (3)
- fannie (3)
- feminism (3)
- fst (3)
- intellectual ventures (3)
- jim simons (3)
- language (3)
- lee kwan yew (3)
- lewontin fallacy (3)
- lhc (3)
- magic (3)
- michael lewis (3)
- mit (3)
- nathan myhrvold (3)
- neal stephenson (3)
- olympiads (3)
- path integrals (3)
- risk preference (3)
- search (3)
- sec (3)
- sivs (3)
- society generale (3)
- systemic risk (3)
- thailand (3)
- twitter (3)
- alibaba (2)
- bear stearns (2)
- bruce springsteen (2)
- charles babbage (2)
- cloning (2)
- david mamet (2)
- digital books (2)
- donald mackenzie (2)
- drugs (2)
- dune (2)
- exchange rates (2)
- frauds (2)
- freddie (2)
- gaussian copula (2)
- heinlein (2)
- industrial revolution (2)
- james watson (2)
- ltcm (2)
- mating (2)
- mba (2)
- mccain (2)
- monkeys (2)
- national character (2)
- nicholas metropolis (2)
- no holds barred (2)
- offices (2)
- oligarchs (2)
- palin (2)
- population structure (2)
- prisoner's dilemma (2)
- singapore (2)
- skidelsky (2)
- socgen (2)
- sprints (2)
- star wars (2)
- ussr (2)
- variance (2)
- virtual reality (2)
- war nerd (2)
- abx (1)
- anathem (1)
- andrew lo (1)
- antikythera mechanism (1)
- athens (1)
- atlas shrugged (1)
- ayn rand (1)
- bay area (1)
- beats (1)
- book search (1)
- bunnie huang (1)
- car dealers (1)
- carlos slim (1)
- catastrophe bonds (1)
- cdos (1)
- ces 2008 (1)
- chance (1)
- children (1)
- cochran-harpending (1)
- cpi (1)
- david x. li (1)
- dick cavett (1)
- dolomites (1)
- eharmony (1)
- eliot spitzer (1)
- escorts (1)
- faces (1)
- fads (1)
- favorite posts (1)
- fiber optic cable (1)
- francis crick (1)
- gary brecher (1)
- gizmos (1)
- greece (1)
- greenspan (1)
- hypocrisy (1)
- igon value (1)
- iit (1)
- inflation (1)
- information asymmetry (1)
- iphone (1)
- jack kerouac (1)
- jaynes (1)
- jazz (1)
- jfk (1)
- john dolan (1)
- john kerry (1)
- john paulson (1)
- john searle (1)
- john tierney (1)
- jonathan littell (1)
- las vegas (1)
- lawyers (1)
- lehman auction (1)
- les bienveillantes (1)
- lowell wood (1)
- lse (1)
- machine (1)
- mcgeorge bundy (1)
- mexico (1)
- michael jackson (1)
- mickey rourke (1)
- migration (1)
- money:tech (1)
- myron scholes (1)
- netwon institute (1)
- networks (1)
- newton institute (1)
- nfl (1)
- oliver stone (1)
- phil gramm (1)
- philanthropy (1)
- philip greenspun (1)
- portfolio theory (1)
- power laws (1)
- pyschology (1)
- randomness (1)
- recession (1)
- sales (1)
- skype (1)
- standard deviation (1)
- starship troopers (1)
- students today (1)
- teleportation (1)
- tierney lab blog (1)
- tomonaga (1)
- tyler cowen (1)
- venice (1)
- violence (1)
- virtual meetings (1)
- wealth effect (1)