This new preprint obtains very interesting results using data from the FinnGen cohort of 300k+ Finns (genotypes + medical records) and UK Biobank.
Genetic risk factors have a substantial impact on healthy life years
Sakari Jukarainen et al.
The impact of genetic variation on overall disease burden has not been comprehensively evaluated. Here we introduce an approach to estimate the effect of different types of genetic risk factors on disease burden quantified through disability-adjusted life years (DALYs, “lost healthy life years”). We use genetic information from 735,748 individuals with registry-based follow-up of up to 48 years. At the individual level, rare variants had higher effects on DALYs than common variants, while common variants were more relevant for population-level disease burden. Among common variants, rs3798220 (LPA) had the strongest effect, with 1.18 DALYs attributable to carrying 1 vs 0 copies of the minor allele. Belonging to top 10% vs bottom 90% of a polygenic score for multisite chronic pain had an effect of 3.63 DALYs. Carrying a deleterious rare variant in LDLR, MYBPC3, or BRCA1/2 had an effect of around 4.1-13.1 DALYs. The population-level disease burden attributable to some common variants is comparable to the burden from modifiable risk factors such as high sodium intake and low physical activity. Genetic risk factors can explain a sizeable number of healthy life years lost both at the individual and population level, highlighting the importance of incorporating genetic information into public health efforts.
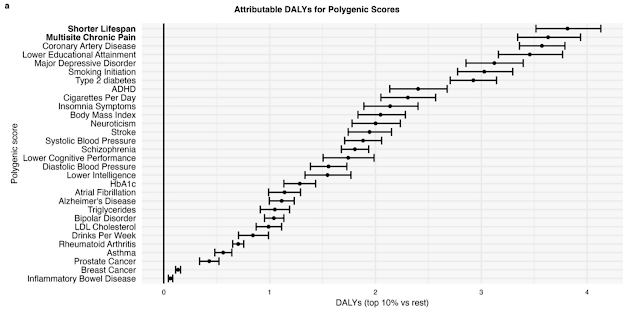
The figure below shows DALYs attributable to being in the top 10% vs
bottom 90% of each PGS. (So, roughly, top 10% vs average individuals.)
The Shorter Lifespan Polygenic Score is a kind of index similar to the Embryo Health Score used by Genomic Prediction. Note that the difference between 90+ percentile and average individuals is roughly 4 DALYs!
In our 2019 sibling validation paper we showed that disease risk polygenic scores have roughly as much predictive power to differentiate high and low risk sibs as when applied to pairs of unrelated individuals.
Thus the results above are indicative of DALY gains from embryo selection.
In 2018 we had Arno Palotie, one of the founders of FinnGen, at MSU to give a talk about the project.


No comments:
Post a Comment